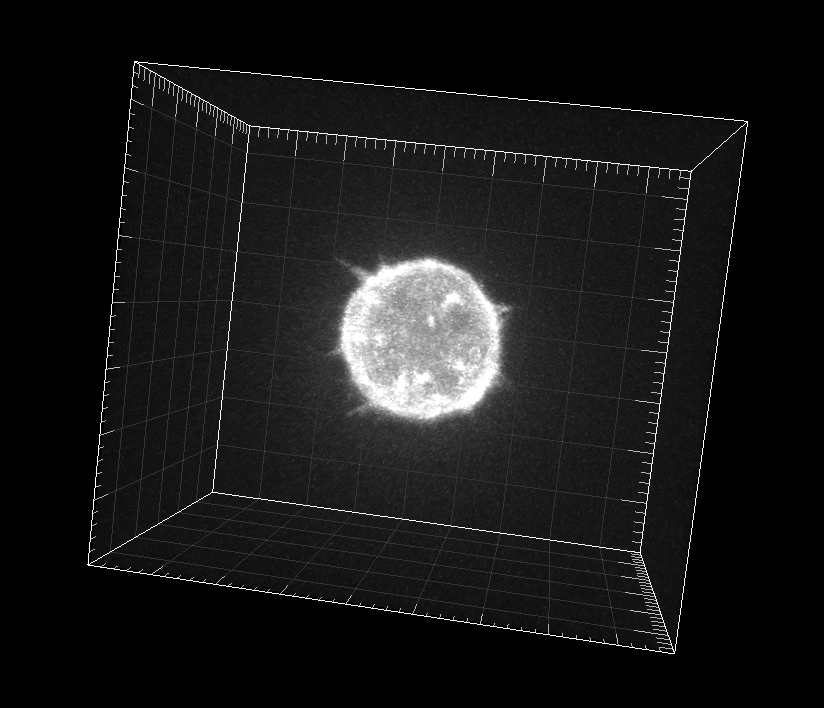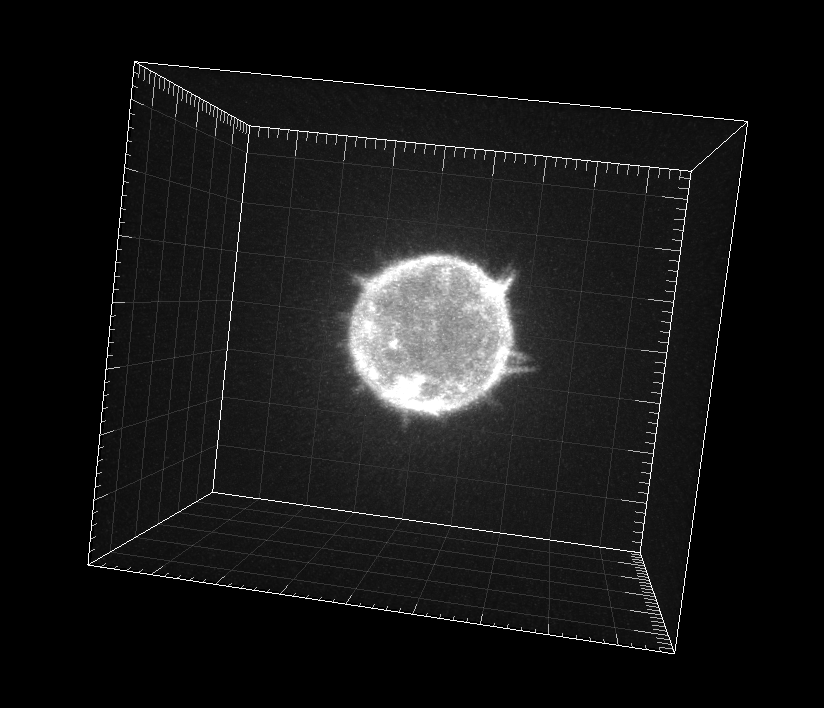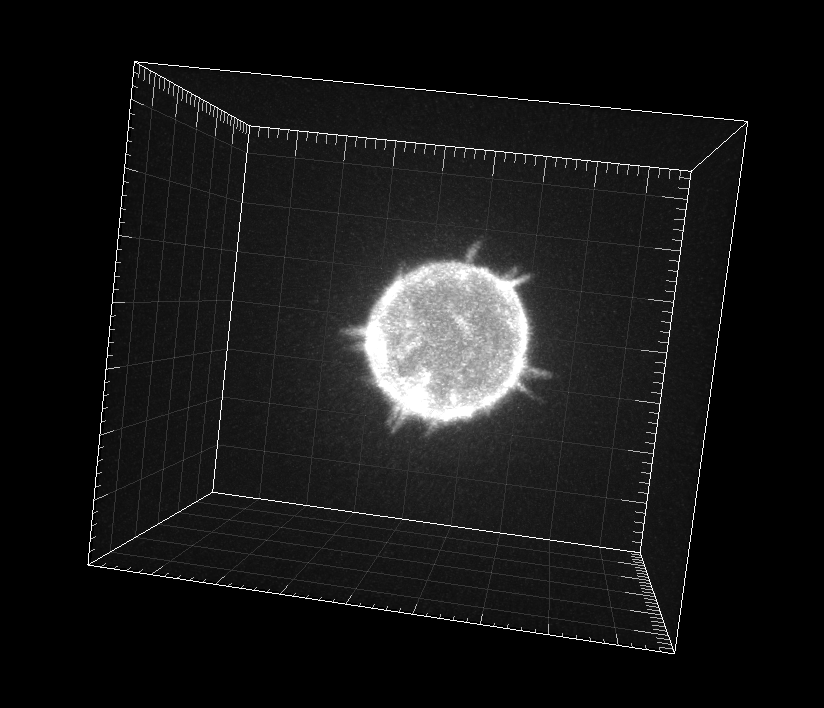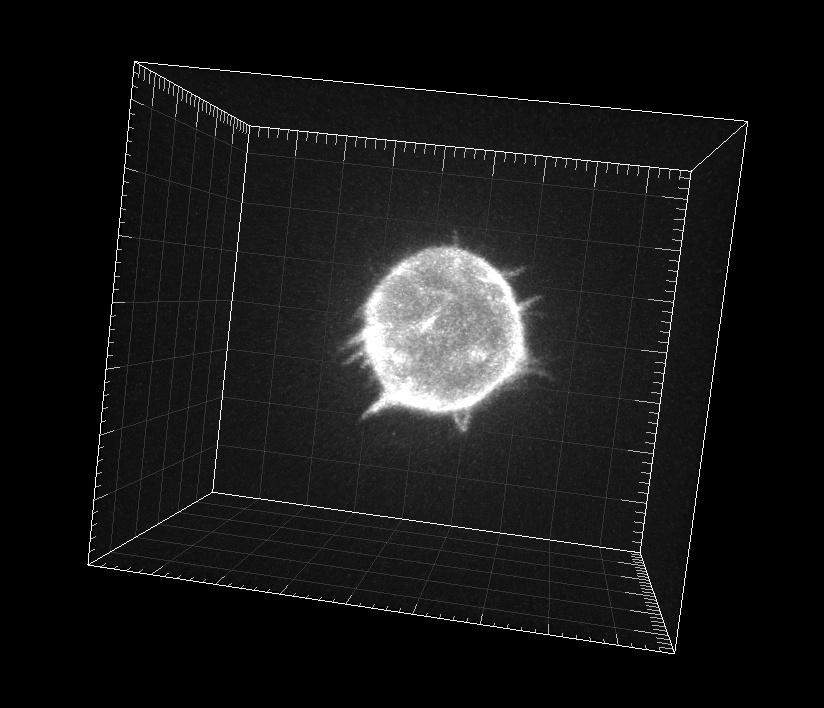
GFP-actin-stained A549 Lung Cancer cells embedded in a Matrigel matrix
Dr. A. Rouzaut. Cell Adhesion and Metastasis Laboratory, Center for Applied Medical Research (CIMA), Pamplona, Spain
Training dataset: http://data.celltrackingchallenge.net/training-datasets/Fluo-C3DH-A549.zip✱ (244 MB)
Test dataset: http://data.celltrackingchallenge.net/test-datasets/Fluo-C3DH-A549.zip (294 MB)

GFP-transfected H157 Lung Cancer cells embedded in a matrigel matrix
Dr. A. Rouzaut. Cell Adhesion and Metastasis Laboratory, Center for Applied Medical Research (CIMA), Pamplona, Spain
Training dataset: http://data.celltrackingchallenge.net/training-datasets/Fluo-C3DH-H157.zip✱ (7.0 GB)
Test dataset: http://data.celltrackingchallenge.net/test-datasets/Fluo-C3DH-H157.zip (7.1 GB)
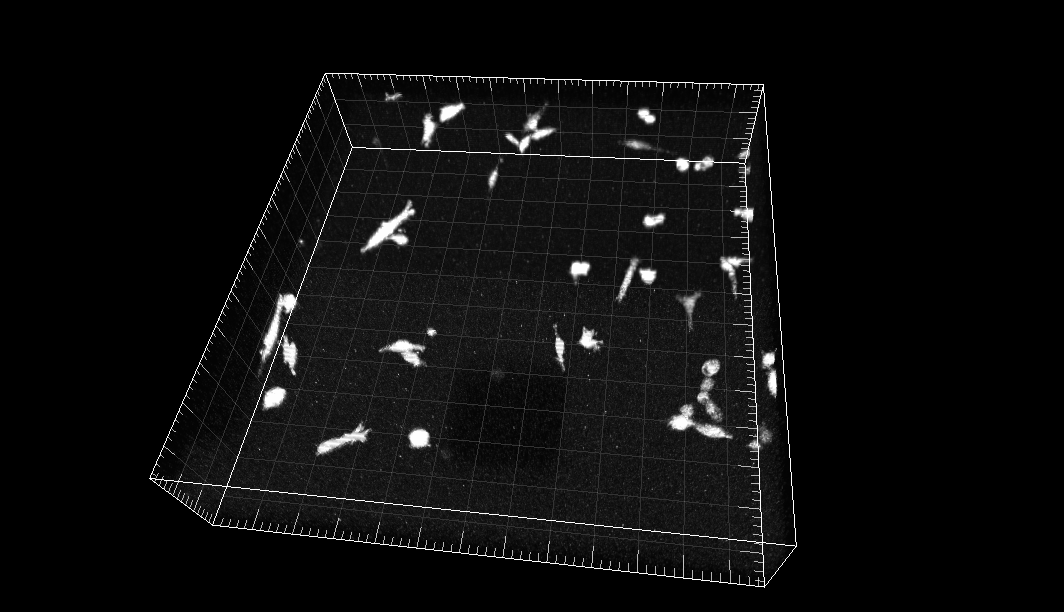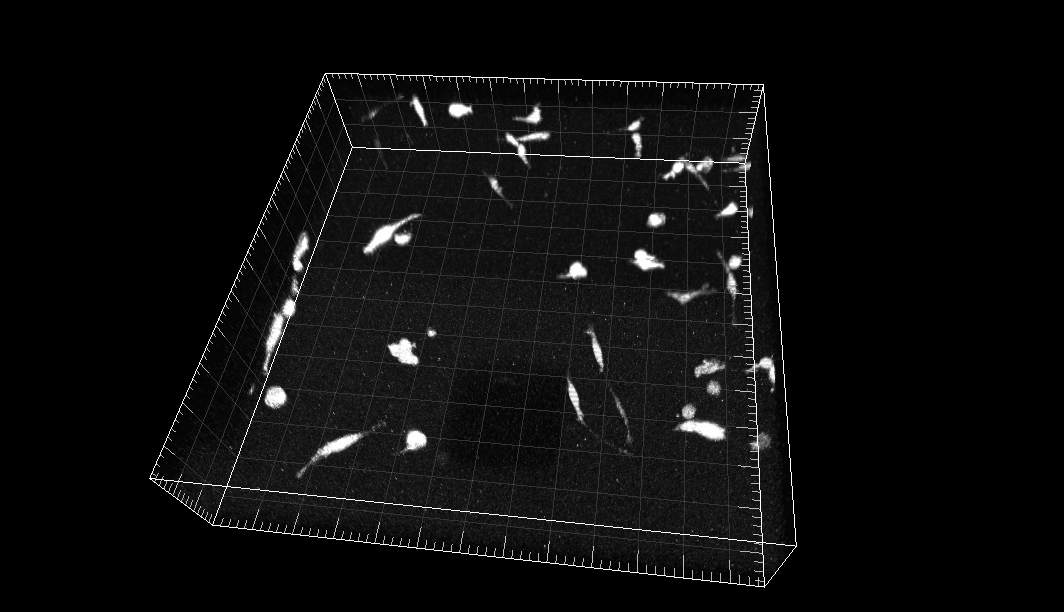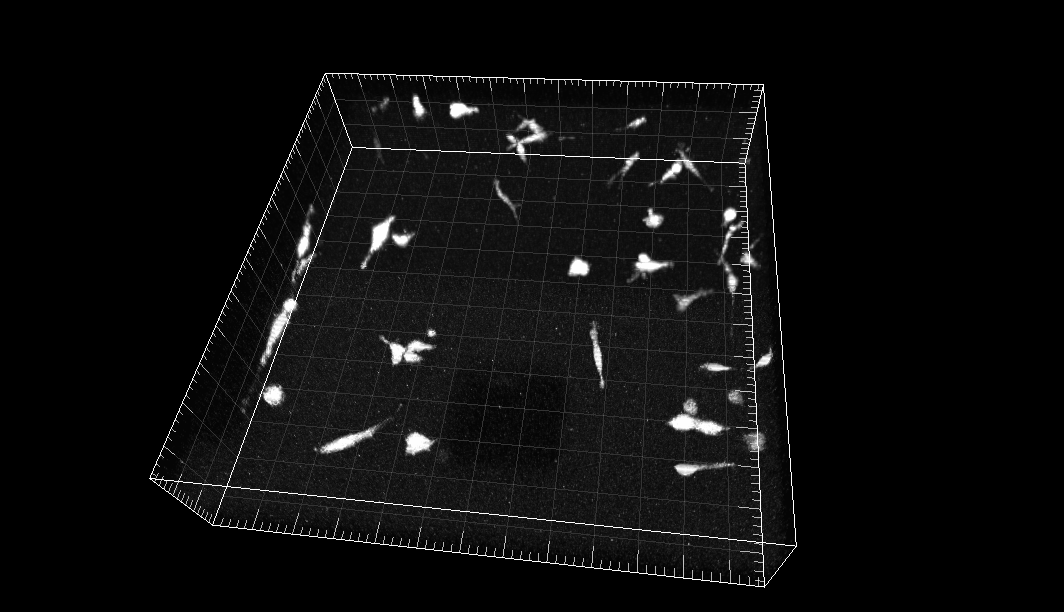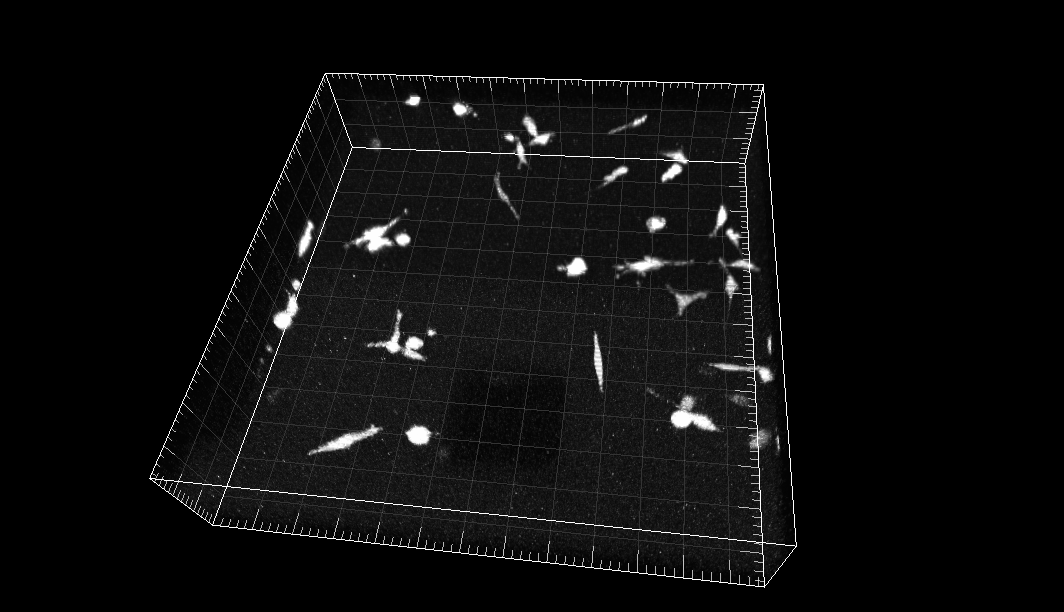
MDA231 human breast carcinoma cells infected with a pMSCV vector including the GFP sequence, embedded in a collagen matrix
Dr. R. Kamm. Dept. of Biological Engineering, Massachusetts Institute of Technology, Cambridge MA (USA)
Training dataset: http://data.celltrackingchallenge.net/training-datasets/Fluo-C3DL-MDA231.zip✱ (182 MB)
Test dataset: http://data.celltrackingchallenge.net/test-datasets/Fluo-C3DL-MDA231.zip (179 MB)
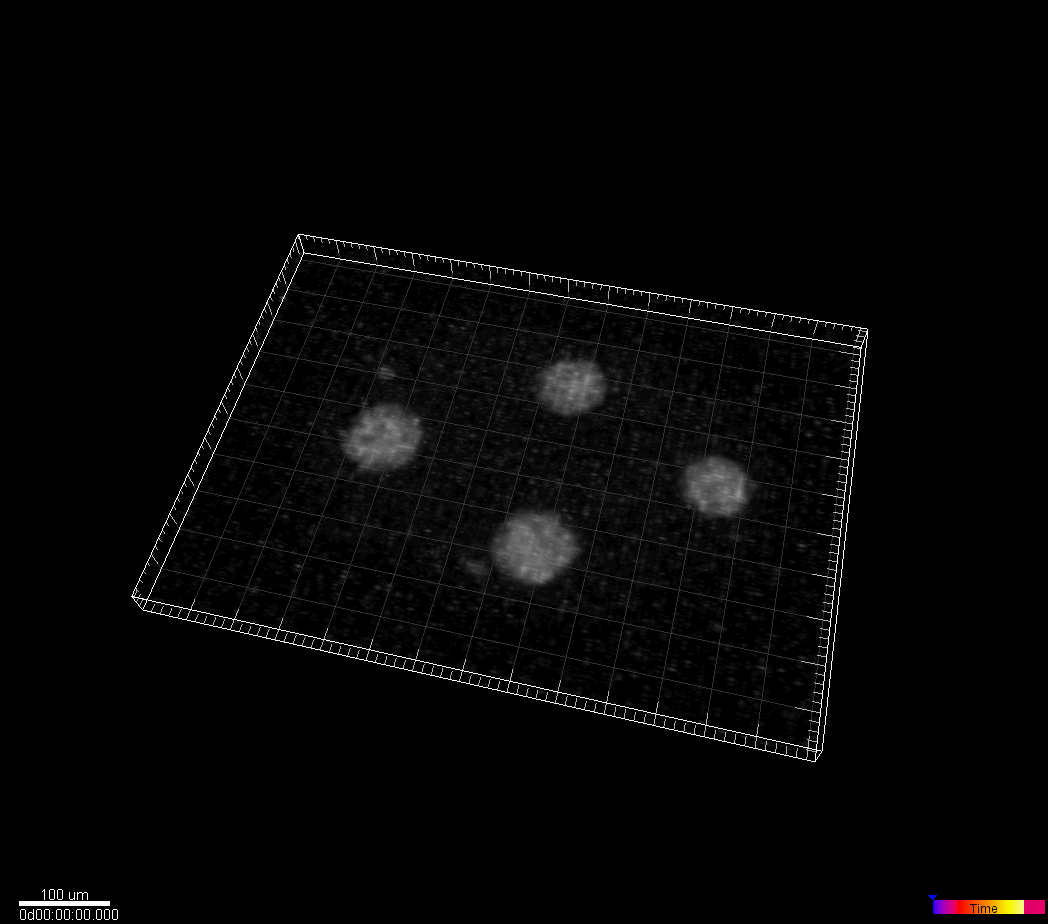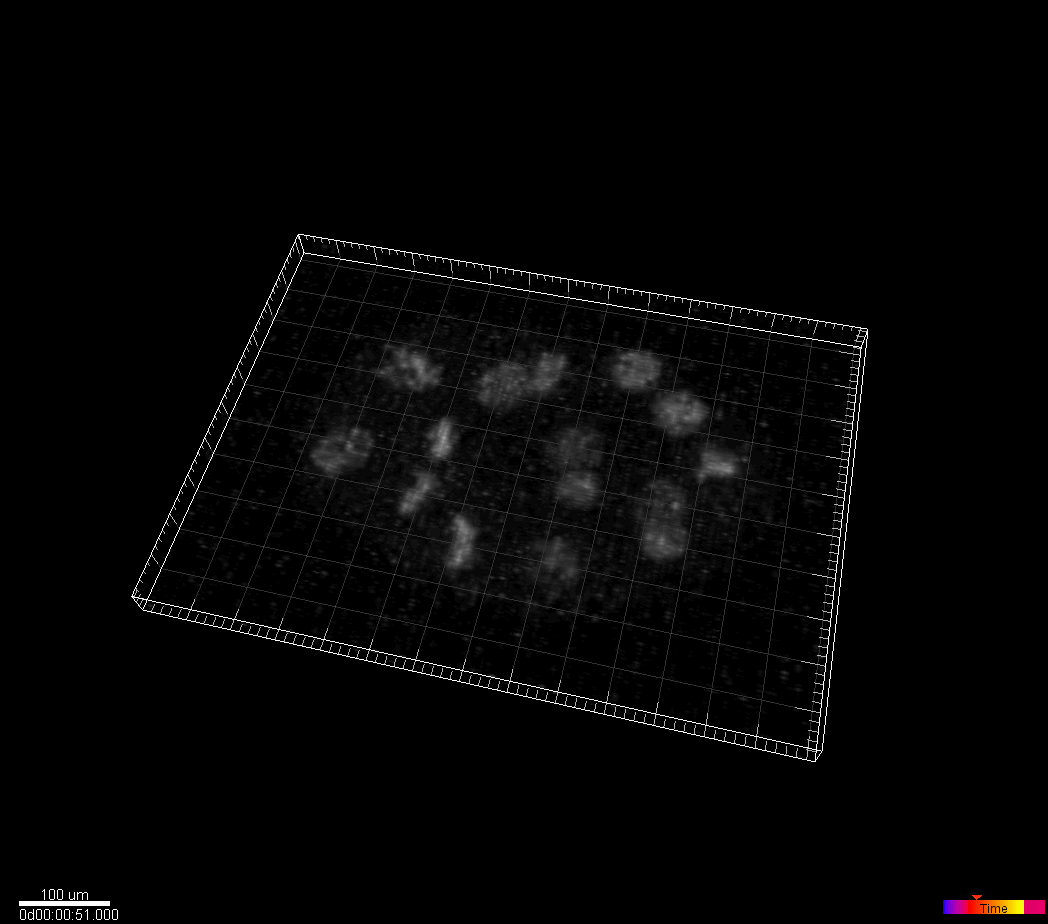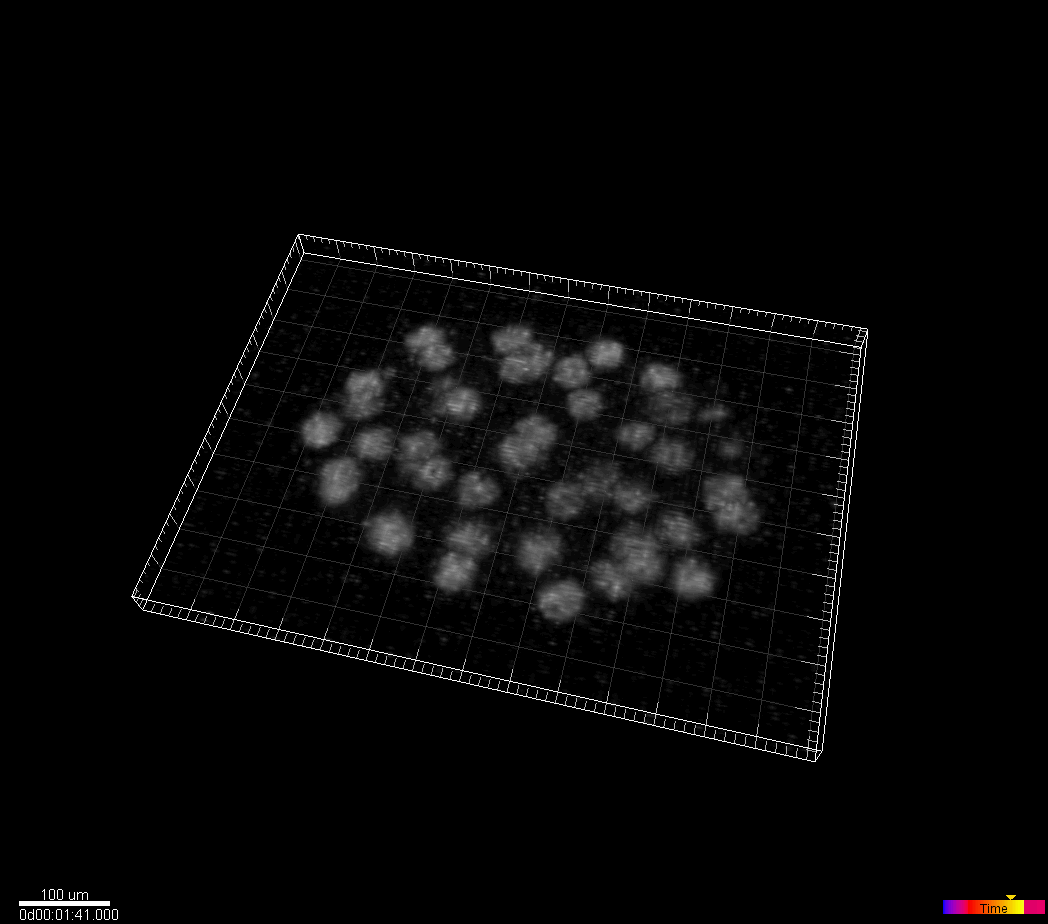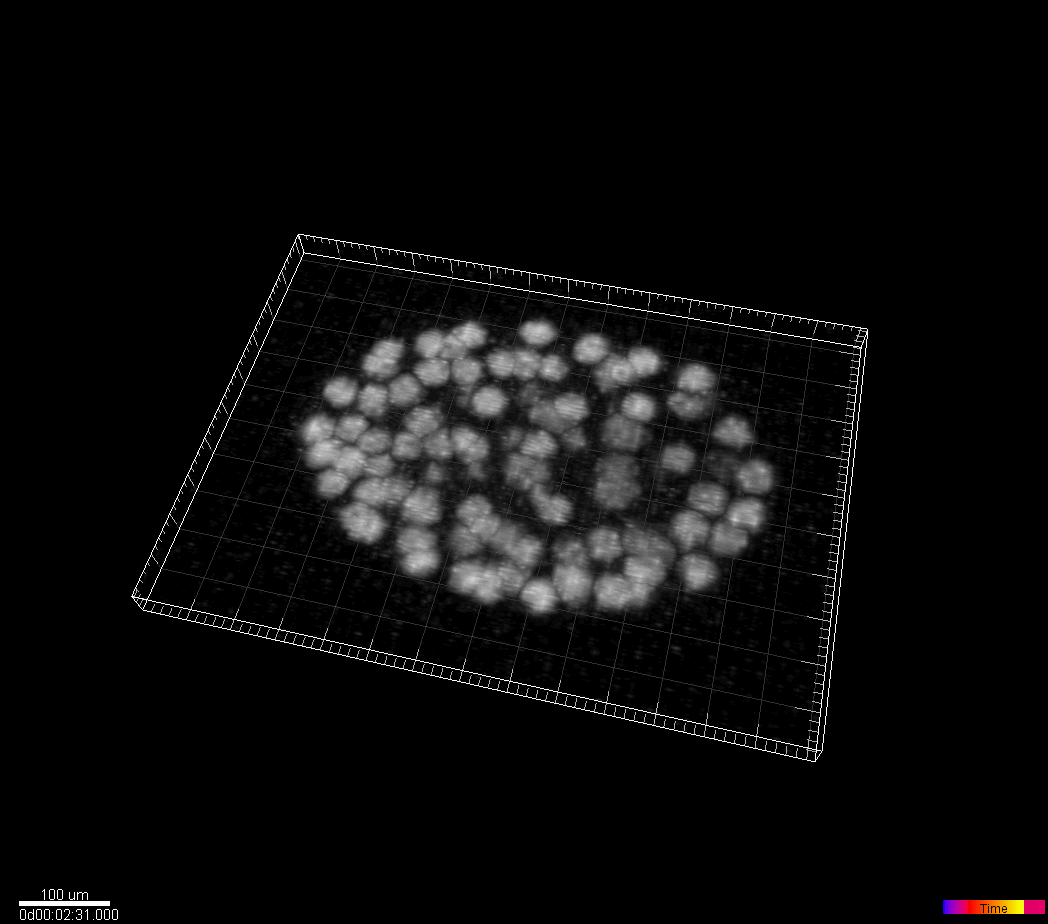
C.elegans developing embryo
Waterston Lab, University of Washington, Seattle, WA, USA
Training dataset: http://data.celltrackingchallenge.net/training-datasets/Fluo-N3DH-CE.zip✱ (3.1 GB)
Test dataset: http://data.celltrackingchallenge.net/test-datasets/Fluo-N3DH-CE.zip (1.7 GB)
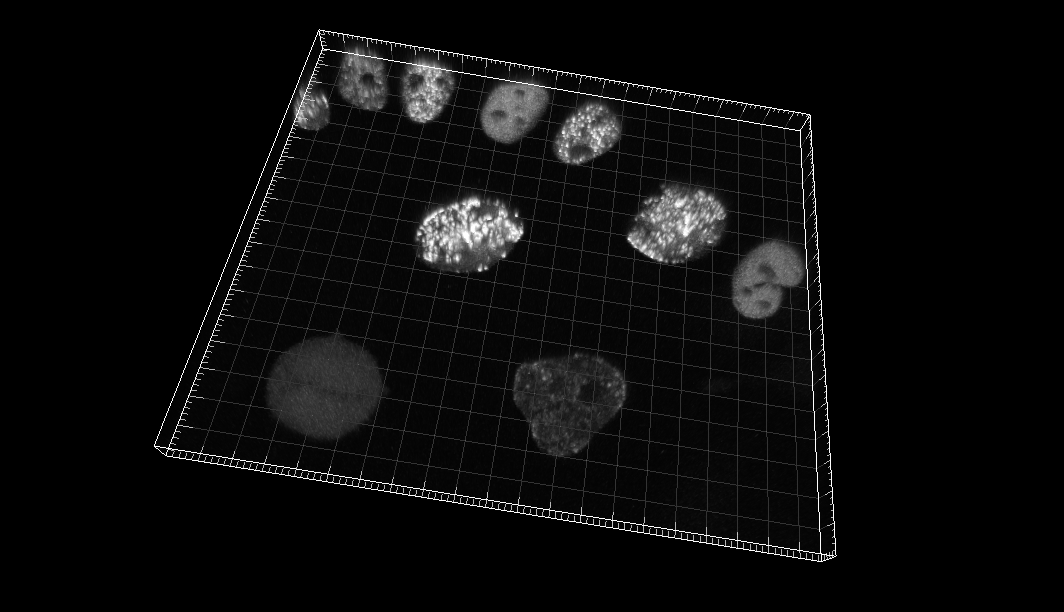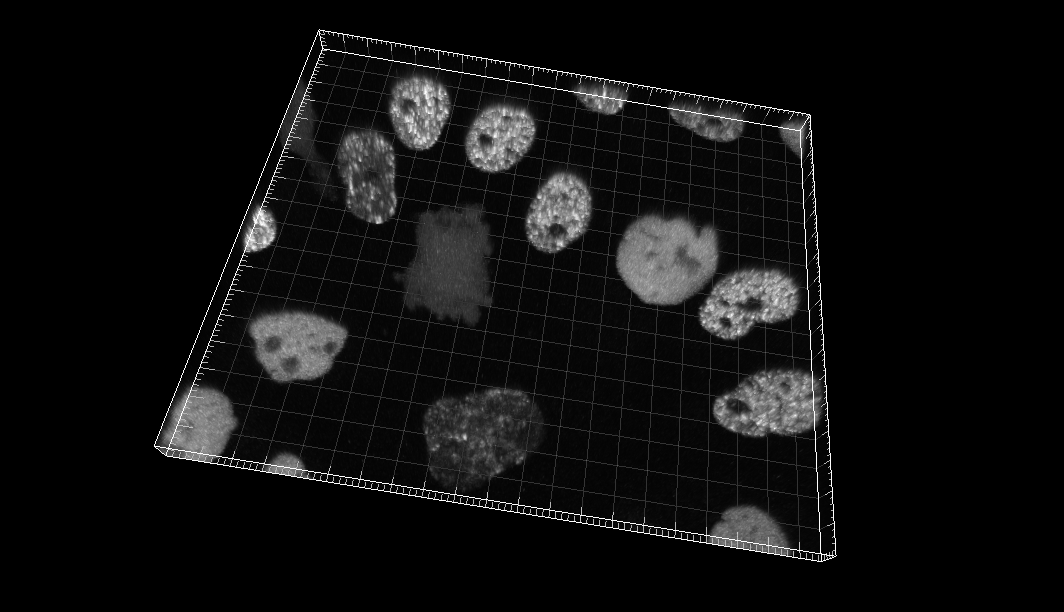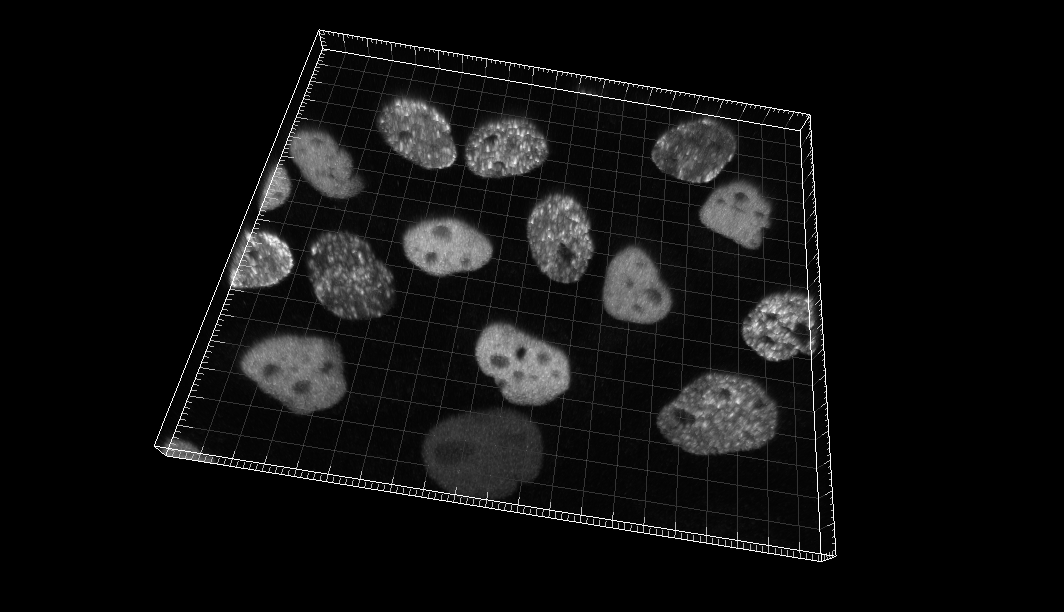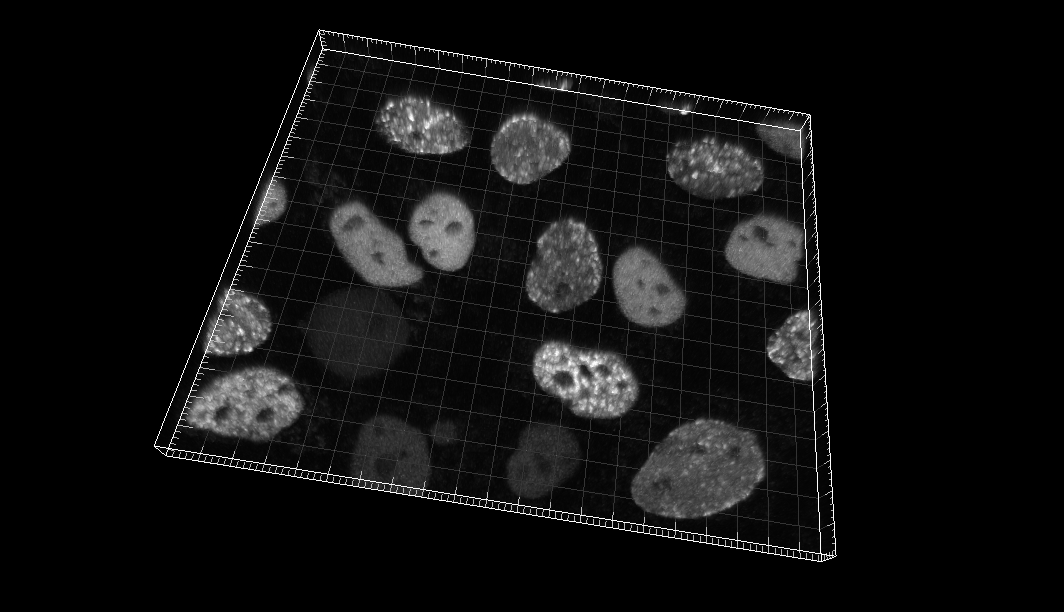
Chinese Hamster Ovarian (CHO) nuclei overexpressing GFP-PCNA
Dr. J. Essers. Dept. of Cell Biology, Erasmus Medical Center, Rotterdam, The Netherlands
Training dataset: http://data.celltrackingchallenge.net/training-datasets/Fluo-N3DH-CHO.zip✱ (98 MB)
Test dataset: http://data.celltrackingchallenge.net/test-datasets/Fluo-N3DH-CHO.zip (105 MB)

Developing Drosophila Melanogaster embryo
Dr. P. Keller. Howard Hughes Medical Institute, Janelia Farms Research Campus, Ashburn VA, USA
Training dataset: http://data.celltrackingchallenge.net/training-datasets/Fluo-N3DL-DRO.zip (5.8 GB)
Test dataset: http://data.celltrackingchallenge.net/test-datasets/Fluo-N3DL-DRO.zip (5.9 GB)

Developing Tribolium Castaneum embryo (3D cartographic projection)
Dr. A. Jain. Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany
Training dataset: http://data.celltrackingchallenge.net/training-datasets/Fluo-N3DL-TRIC.zip (20.6 GB)
Test dataset: http://data.celltrackingchallenge.net/test-datasets/Fluo-N3DL-TRIC.zip (19.9 GB)

Developing Tribolium Castaneum embryo
Dr. A. Jain. Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany
Training dataset: http://data.celltrackingchallenge.net/training-datasets/Fluo-N3DL-TRIF.zip (320 GB)
Test dataset: http://data.celltrackingchallenge.net/test-datasets/Fluo-N3DL-TRIF.zip (467 GB)
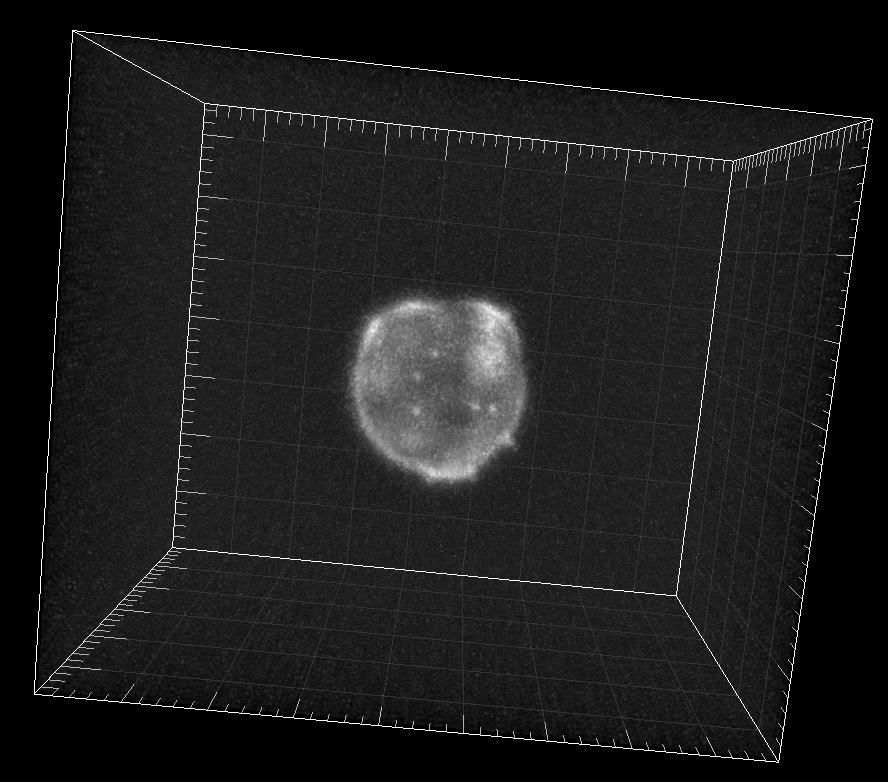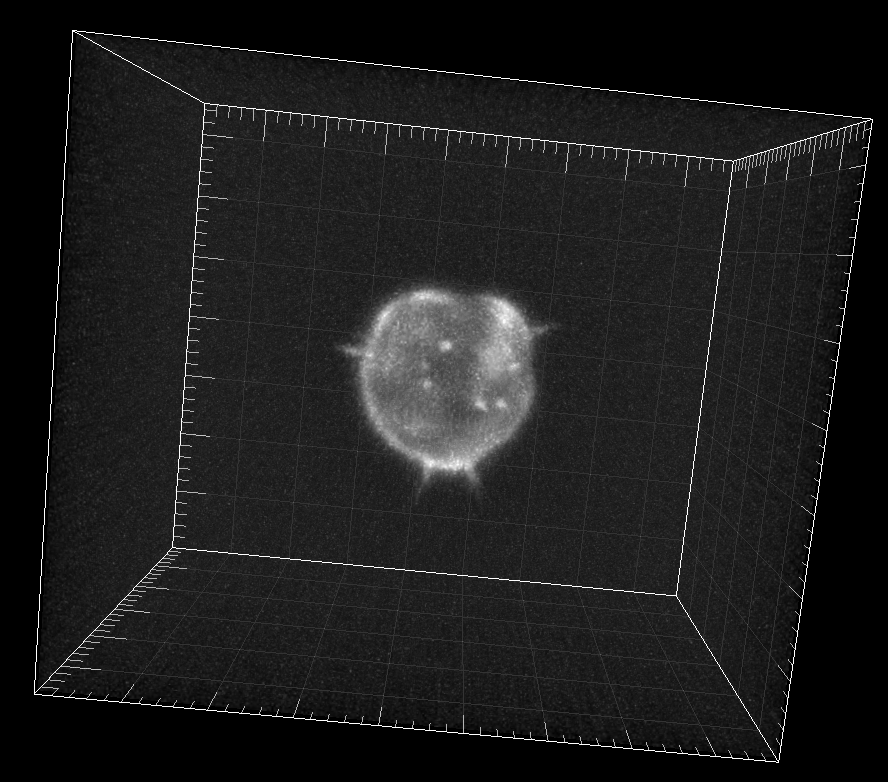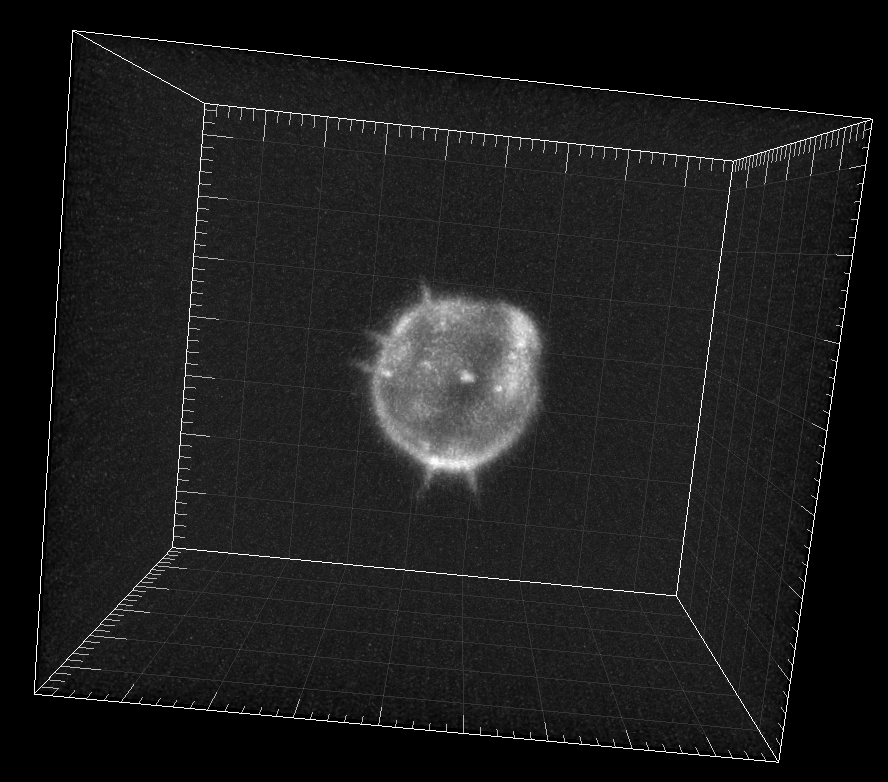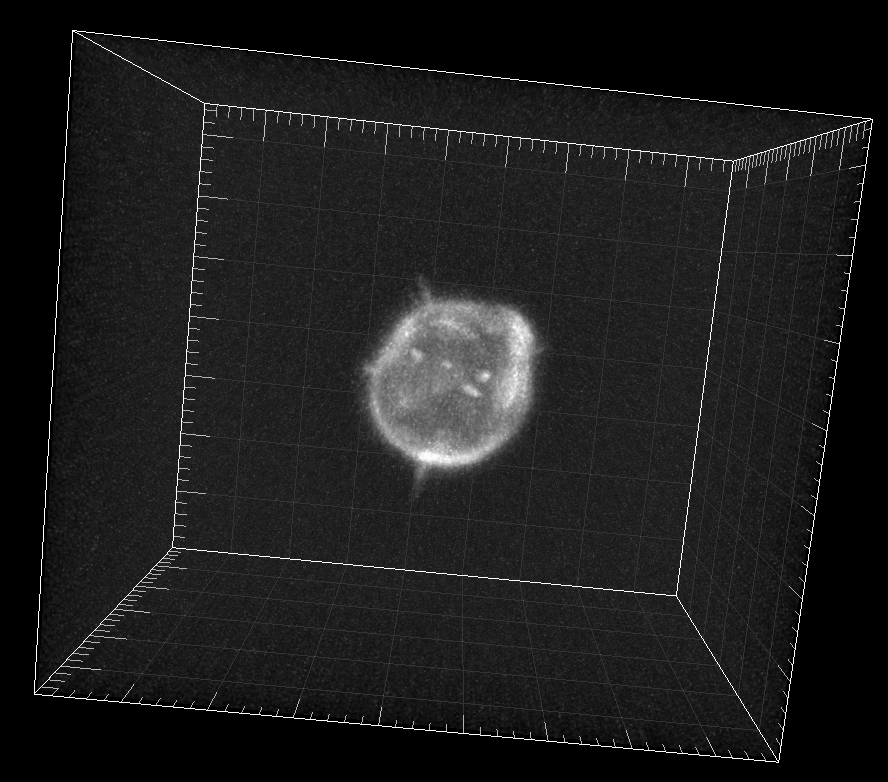
Simulated GFP-actin-stained A549 Lung Cancer cells embedded in a Matrigel matrix
Dr. M. Maška and Dr. D. V. Sorokin. Centre for Biomedical Image Analysis (CBIA),
Masaryk University, Brno, Czech Republic (Created using FiloGen, part of Cytopacq)
Training dataset: http://data.celltrackingchallenge.net/training-datasets/Fluo-C3DH-A549-SIM.zip✚ (314 MB)
Test dataset: http://data.celltrackingchallenge.net/test-datasets/Fluo-C3DH-A549-SIM.zip (327 MB)
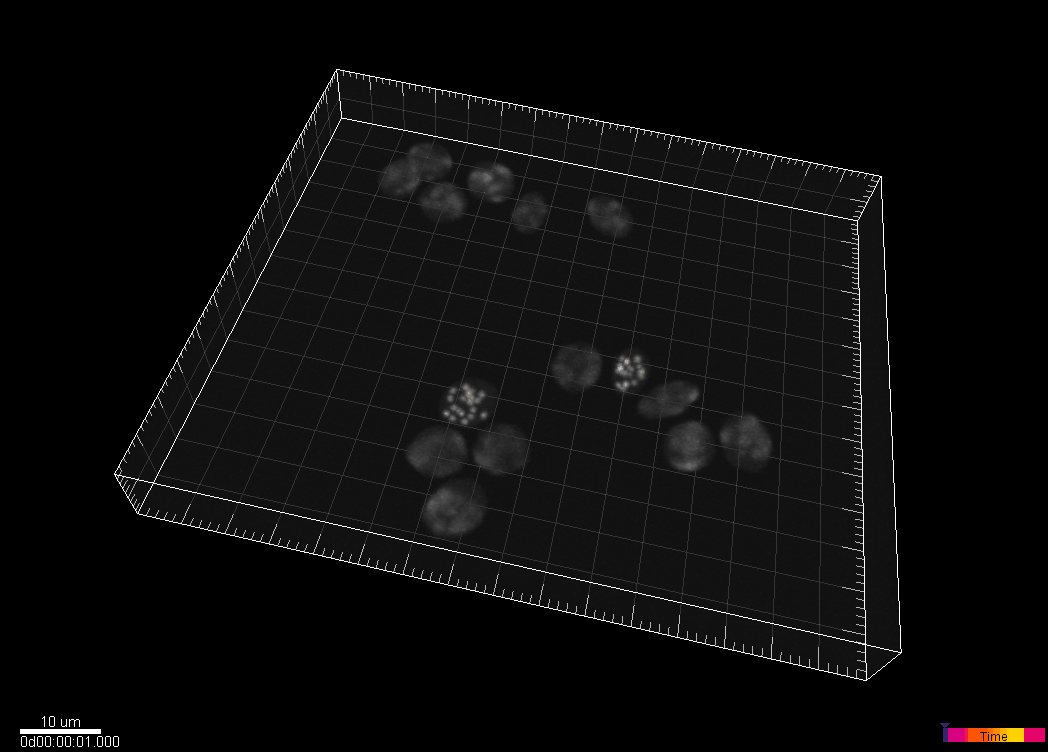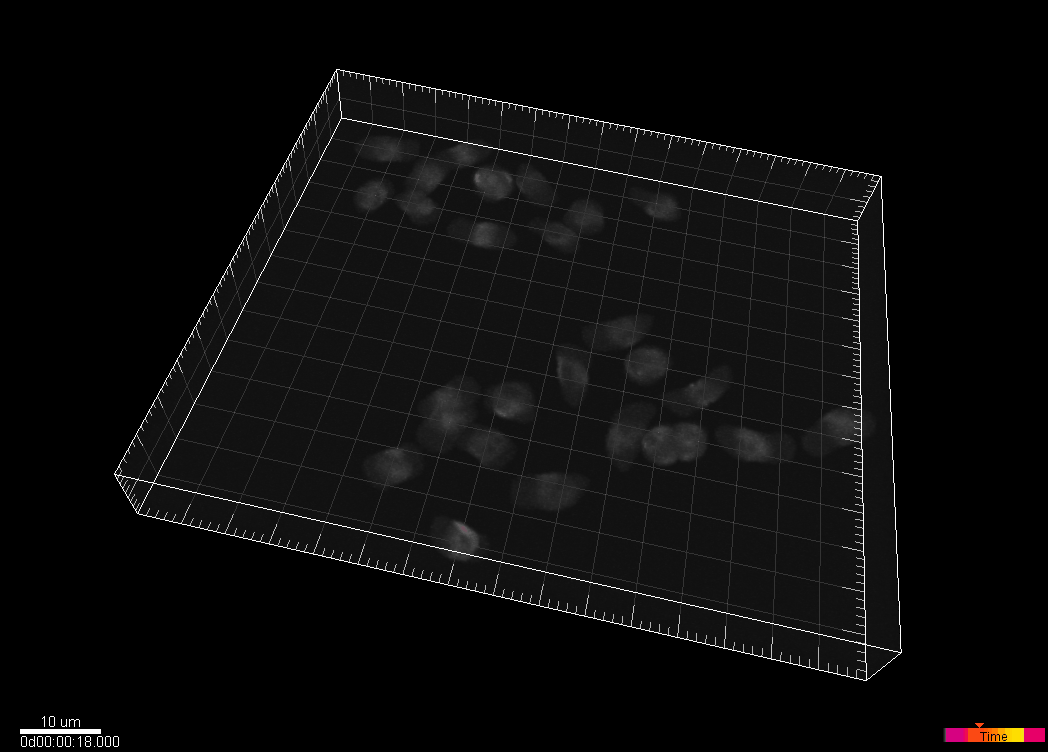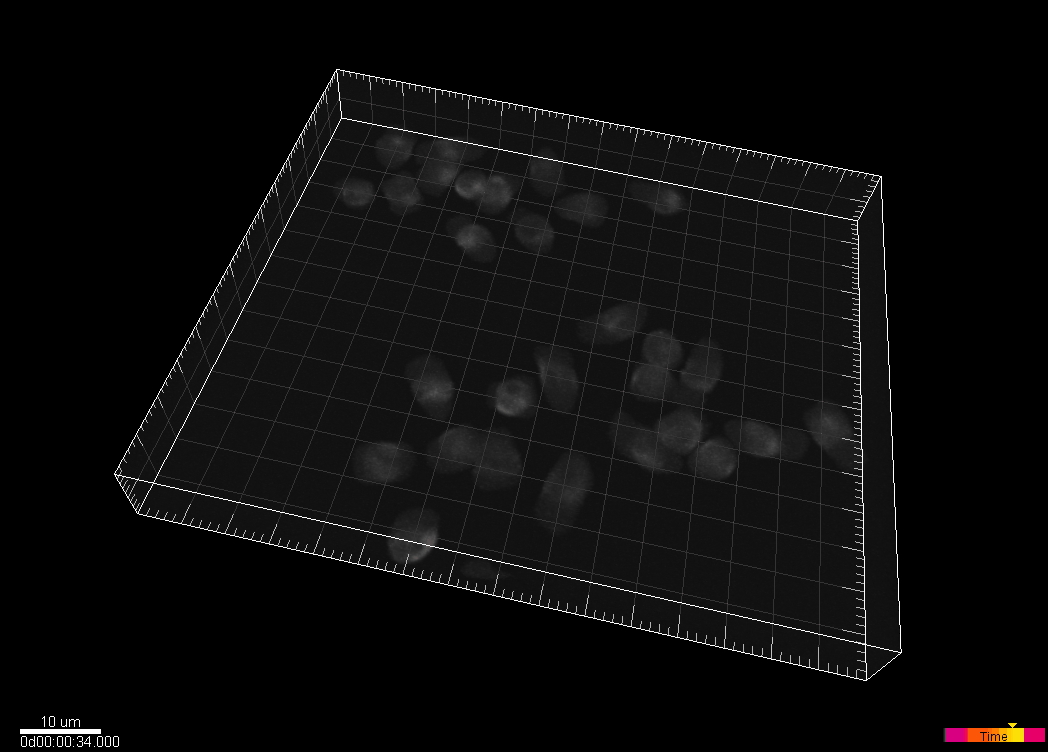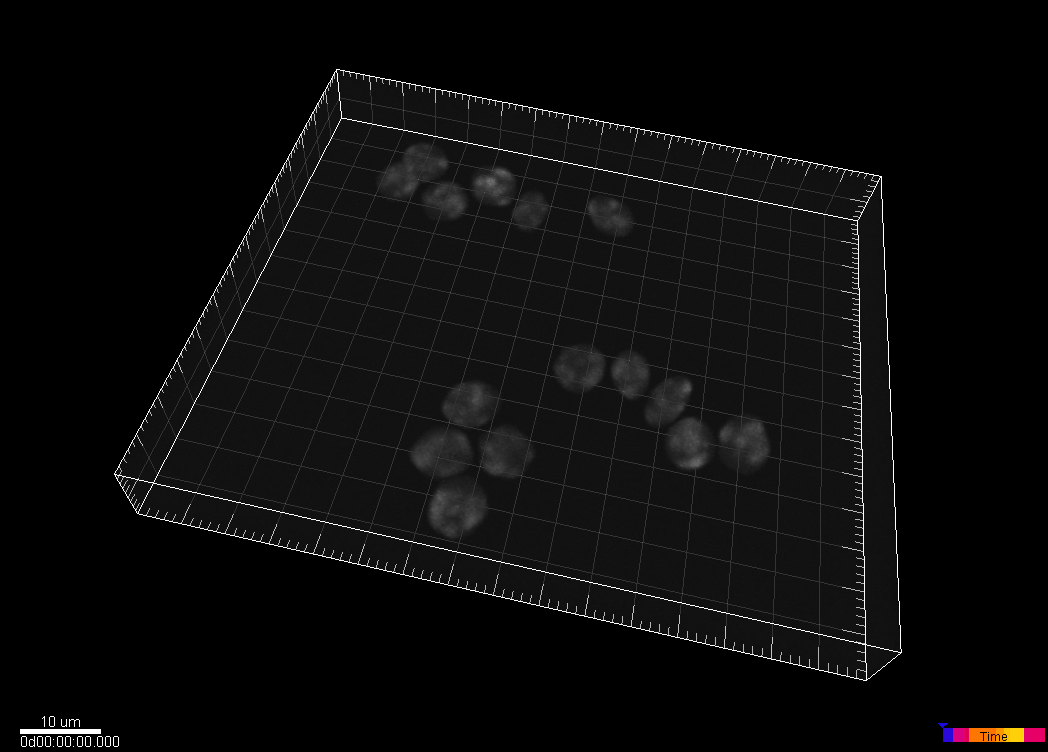
Simulated nuclei of HL60 cells stained with Hoechst
Dr. V. Ulman and Dr. D. Svoboda. Centre for Biomedical Image Analysis (CBIA),
Masaryk University, Brno, Czech Republic (Created using MitoGen, part of Cytopacq)
Training dataset: http://data.celltrackingchallenge.net/training-datasets/Fluo-N3DH-SIM+.zip✚ (3.1 GB)
Test dataset: http://data.celltrackingchallenge.net/test-datasets/Fluo-N3DH-SIM+.zip (5.9 GB)
✱ The silver reference segmentation annotations and imperfect segmentation masks to link are available for a particular real training dataset.
✚ The perfect segmentation masks to link are available for a particular computer-generated training dataset.