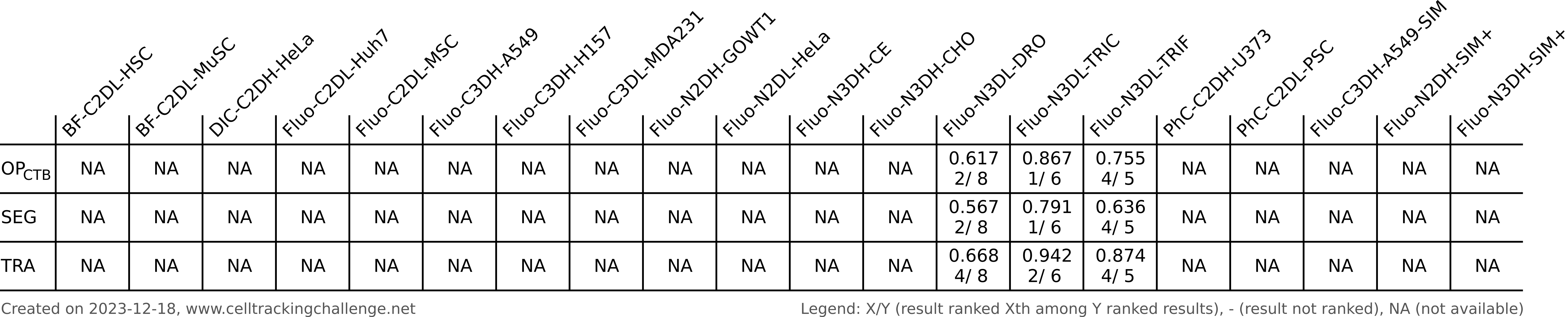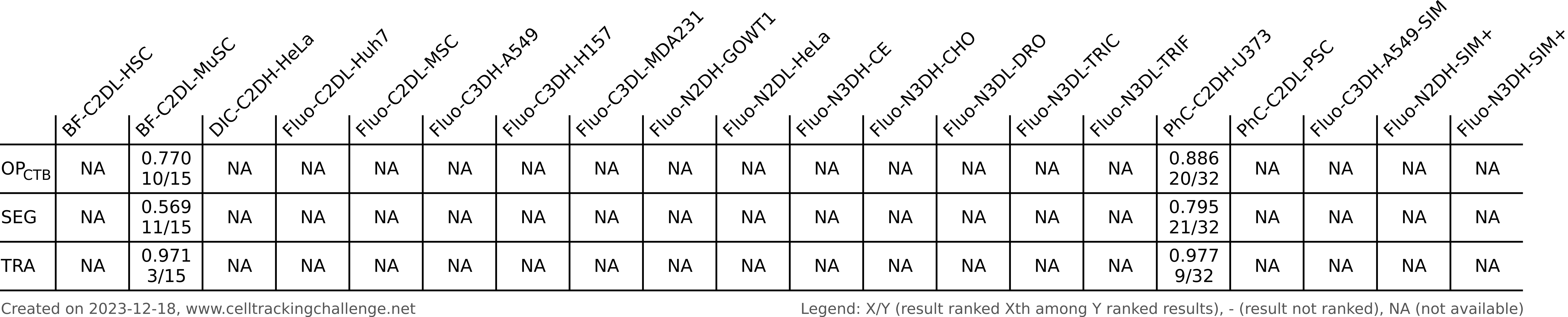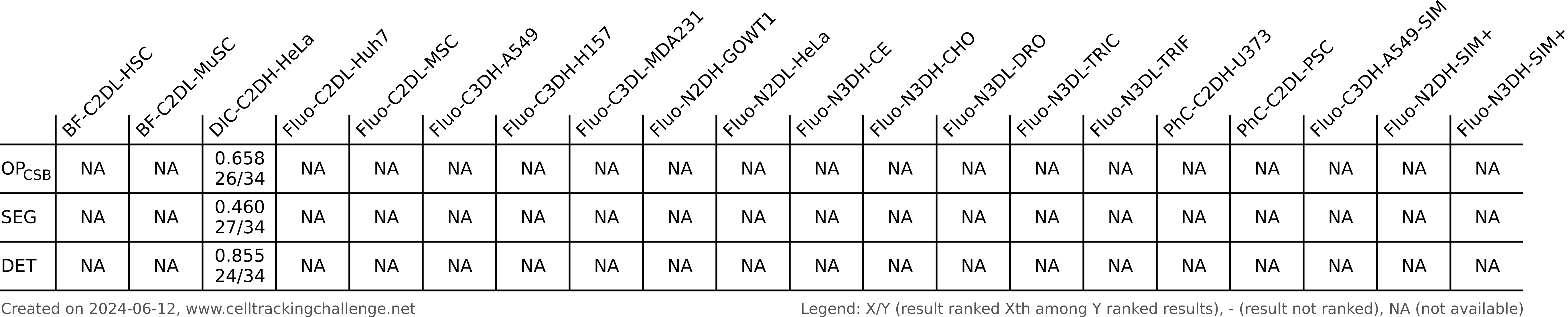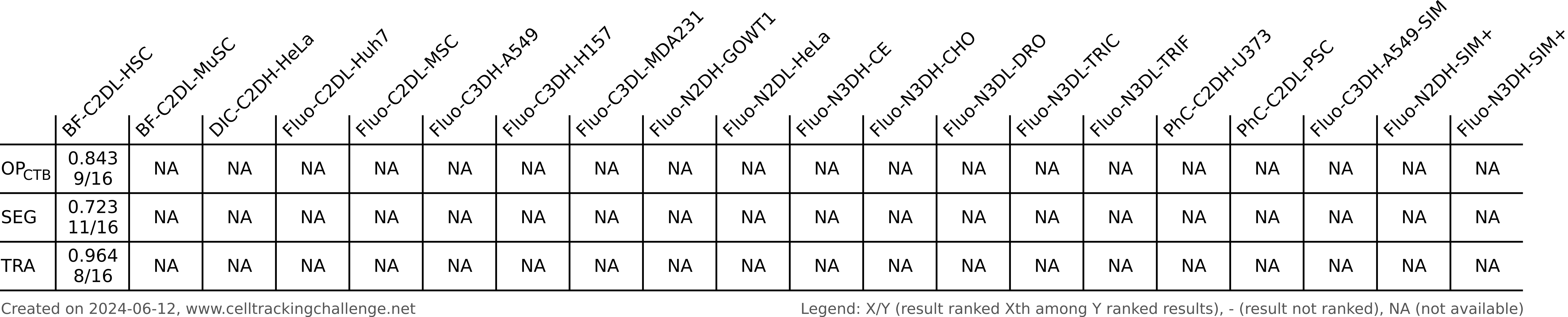Authors
Klas Magnusson – since 2017: RaySearch Laboratories, Stockholm, Sweden
until 2016: KTH Royal Institute of Technology, ACCESS Linnaeus Center, Stockholm, Sweden
Joakim Jaldén – KTH Royal Institute of Technology, ACCESS Linnaeus Center, Stockholm, Sweden
Helen M. Blau – Baxter Laboratory for Stem Cell Biology, Stanford University School of Medicine, Stanford, CA, USA
Contact
Klas Magnusson (klasmagnus@gmail.com)
Algorithms
Additional information
The complete software, including graphical user interfaces for setting of parameters, visualization of tracking results, manual correction and plotting of results can be downloaded from http://github.com/klasma/BaxterAlgorithms. Since the challenge submission, bugs might have been fixed and improvements might have been made, and therefore the tracking results may not be exactly the same as for the executables used in the challenge and available above.